Sudheer Sahu, Peng Yin, & John Reif
In Proc. Eleventh International Meeting on DNA Computing, 2005, to appear
Downloads: PDF
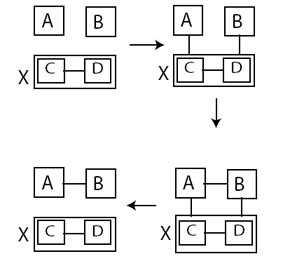
Abstract: We propose a reversible self-assembly model in
which the glue strength between two juxtaposed tiles is a function of the
time they have been in neighboring positions. We then present an implementation
of our model using strand displacement reactions on DNA tiles. Under our model,
we can for the first time demonstrate and study catalysis and self-replication
in the tile assembly rigorously. We then study the tile-complexity for assembling
various shapes in our model. We show that using $O(\frac{\log N}{\log \log N})$ types of tiles, we can assemble thin
rectangles of size $k\times N$. We also show a $\Omega(k^{(N-k)/2})$
lower bound for tile-complexity of a square of size $N\times N$ with an hole
of size $k\times k$ in \emph{Tile Assembly Model} \cite{Rothemund00}, and
an upper bound of $O(\frac{\log N}{\log \log N})$ for the same shape
in our mode.

Back to pulications