Chenxiang Yin, Ralf Jungmann, Andrew M. Leifer, Chao Li, Daniel Levner, George M. Church, William Shih, Peng Yin
Nat Chem. doi: 10.1038/nchem, 832-839 (2012).
Downloads: PDF, 8 pages.
Abstract:
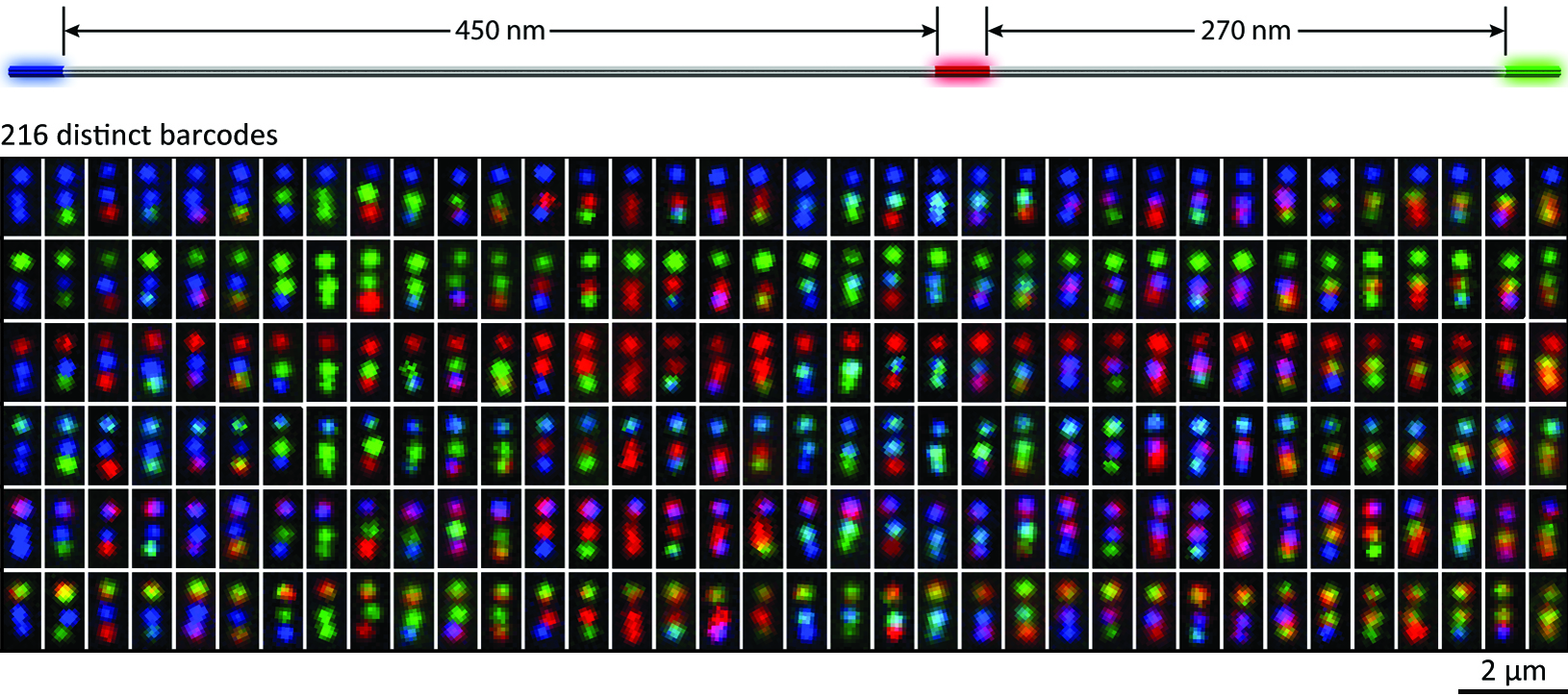
The identification and differentiation of a large number of distinct molecular species with high temporal and spatial resolution is a major challenge in biomedical science. Fluorescence microscopy is a powerful tool, but its multiplexing ability is limited by the number of spectrally distinguishable fluorophores. Here, we used (deoxy)ribonucleic acid (DNA)-origami technology to construct submicrometre nanorods that act as fluorescent barcodes. We demonstrate that spatial control over the positioning of fluorophores on the surface of a stiff DNA nanorod can produce 216 distinct barcodes that can be decoded unambiguously using epifluorescence or total internal reflection fluorescence microscopy. Barcodes with higher spatial information density were demonstrated via the construction of super-resolution barcodes with features spaced by -40 nm. One species of the barcodes was used to tag yeast surface receptors, which suggests their potential applications as in situ imaging probes for diverse biomolecular and cellular entities in their native environments.
Supplementary material:PDF, 40 pages

Summary figure: Life-science research and biomedical diagnostics call for robust fluorescence barcodes of compact size and high multiplexing capability. Here DNA-origami technology was used to construct a new kind of geometrically encoded barcodes with excellent structural stiffness. They hold promise for ex and in situ imaging of diverse biologically relevant entities.
Back to publications